Fast-forward on Your Research with EditCo's Reliable CRISPR Cell Line Development Services
EditCo's Cell Line Development Services
.webp?width=2000&height=1500&name=updated_workcell%20(1).webp)
Higher-Quality CRISPR Reagents
Consistent, Quality CRISPR Reagents at Unparalleled Speeds
We are here to accelerate answers to your critical research by providing access to reproducible and high-quality CRISPR gRNAs at unparalleled speeds, through the use of a robust automated inventory management and fulfillment platform.
Faster Turnaround Times
Robust automation allows for delivery of CRISPR reagents in as fast as 5 days, enabling discoveries at the speed of research.
Scalable Precision
Strict quality control combined with intelligent design enables precision high-throughput experiments to fit today's scientific landscape.
Testimonials consectitur amet sed
Congue fermentum eros leo commodo. Integer vitae vitae ac amet. Vitae sed lectus vel lacus adipiscing nulla facilisis mauris porttitor. Viverra elementum odio vitae id.

Viverra aliquam volutpat interdum quisque vel mattis. Malesuada aliquam egestas pharetra a tempus ullamcorper egestas.

Nibh luctus pulvinar sagittis faucibus commodo vitae purus. Ullamcorper dictum.

Congue fermentum eros leo commodo. Integer vitae vitae ac amet. Vitae sed lectus vel lacus adipiscing nulla facilisis mauris porttitor. Viverra elementum odio vitae id.

Congue fermentum eros leo commodo. Integer vitae vitae ac amet. Vitae sed lectus vel lacus adipiscing nulla facilisis mauris porttitor. Viverra elementum odio vitae id.

Congue fermentum eros leo commodo. Integer vitae vitae ac amet. Vitae sed lectus vel lacus adipiscing nulla facilisis mauris porttitor. Viverra elementum odio vitae id.


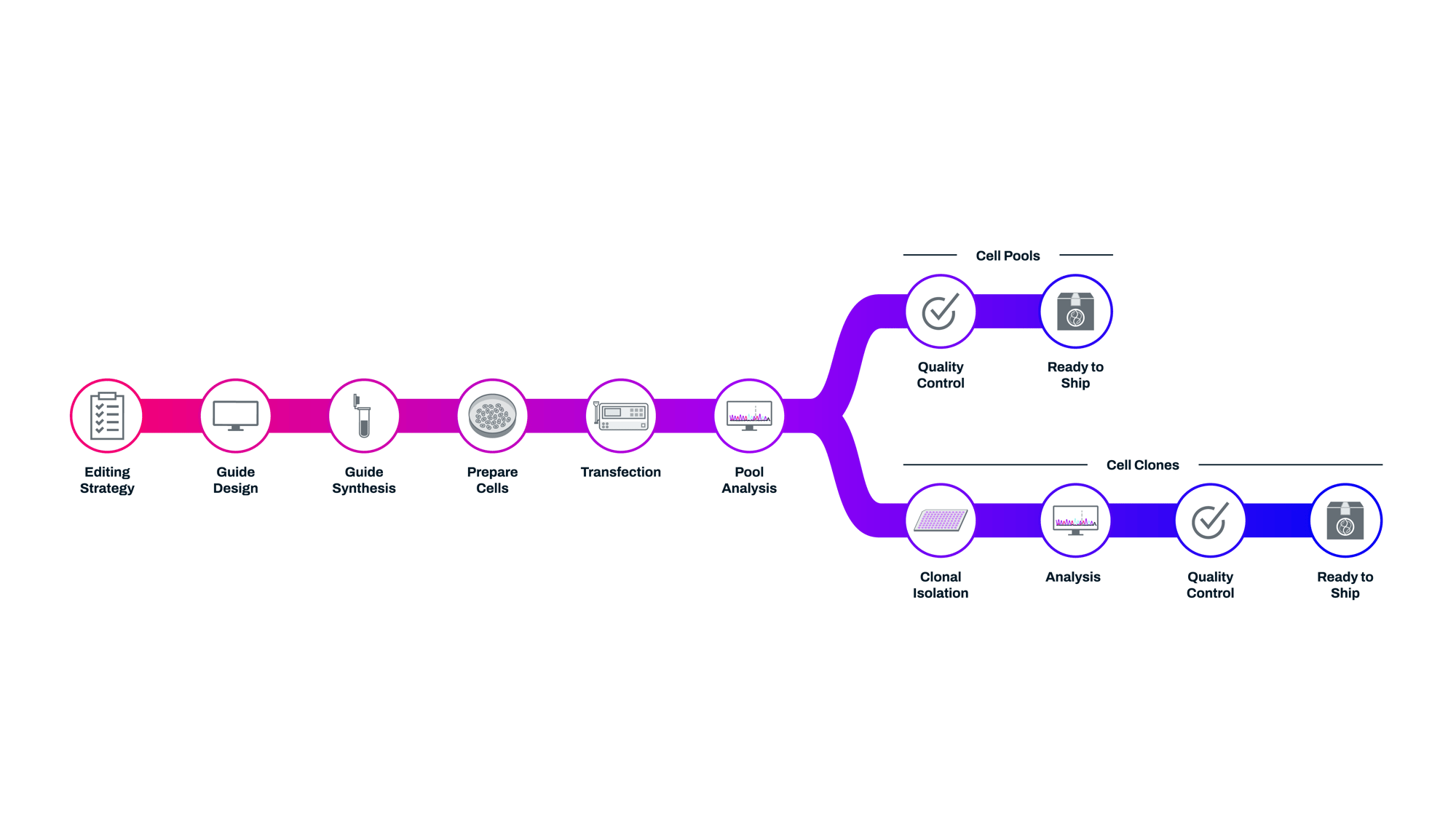
Flexible and efficient CRISPR cell line development through deep EditCo team expertise, highly specialized bioinformatics and automated processes.

White glove service with consistent communication on your timeline through our knowledgable team. EditCo's PhD-level service and commercial teams provide cell models without the need to optimize and troubleshoot to get to your downstream assays.

Precision editing, meticulous tracking, and stingent quality control are a big part of what allows EditCo to build cell models for a vast array of different applications. From immunology, to neuroscience, to infectious disease and beyond, we understand the need for getting the edited cell lines required for your downstream discoveries.
EditCo's CRISPR Platform in Action
.webp?width=2000&height=1500&name=updated_workcell%20(1).webp)