Assay Development
Revolutionizing Assay Development with CRISPR: Towards Greater Accuracy and Efficiency
In the rapidly evolving fields of biotechnology and molecular biology, assay development plays a pivotal role. Assays are analytical procedures that detect, quantify, or characterize the presence or activity of a target molecule, cell, or organism. From drug discovery to disease diagnostics, assays are essential tools for advancing scientific research and medical practice. Traditionally, assay development has relied on established biochemical techniques, but the advent of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology is revolutionizing this field, enabling the creation of more accurate, efficient, and versatile assays.
.png?width=700&name=unnamed%20(75).png)
Fig 1. Cell viability assay using Calcein AM to visualize live cells stained in green and EthD-1 to visualize the nuclei of dead cells stained in red. (Credit: In Vitro Growth of Mouse Preantral Follicles Under Simulated Microgravity, Journal of Visualized Experiments)
The Basics of Assay Development
Assay development involves several key steps, including the identification of a target, selection of appropriate detection methods, optimization of conditions for specificity and sensitivity, and validation of the assay’s reliability. Depending on the application, assays can range from simple colorimetric tests to highly sophisticated techniques involving fluorescence, luminescence, or mass spectrometry. The ultimate goal is to create an assay that is both sensitive (able to detect low levels of the target) and specific (able to distinguish the target from other substances), while also being reproducible, scalable, and cost-effective.
The Role of CRISPR in Assay Development
CRISPR technology, originally discovered as a bacterial immune defense mechanism, has rapidly become a game-changer in genetic engineering and molecular biology. The most commonly used CRISPR system, CRISPR-Cas9, allows researchers to make precise edits to DNA by targeting specific sequences with a guide RNA (gRNA) and using the Cas9 enzyme to introduce cuts. However, beyond its well-known role in gene editing, CRISPR has also shown immense potential in the development of innovative assays.
CRISPR-Based Diagnostics
One of the most exciting applications of CRISPR in assay development is in diagnostics. Traditional diagnostic assays often rely on techniques like polymerase chain reaction (PCR) or enzyme-linked immunosorbent assays (ELISA), which, while effective, can be time-consuming, expensive, and require specialized equipment. CRISPR-based diagnostics offer a faster, more flexible alternative.
For example, CRISPR-Cas12 and Cas13 enzymes have been adapted for diagnostic purposes. Unlike Cas9, which cuts DNA, Cas12 and Cas13 are capable of collateral cleavage, where they indiscriminately cut single-stranded DNA or RNA once activated by a specific target sequence. This property has been harnessed to create highly sensitive and specific assays for detecting viral infections, such as COVID-19, through the SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing) and DETECTR (DNA Endonuclease Targeted CRISPR Trans Reporter) platforms. These CRISPR-based systems can detect the presence of viral RNA or DNA with a simple color change or fluorescence signal, allowing for rapid, point-of-care testing without the need for complex laboratory infrastructure.
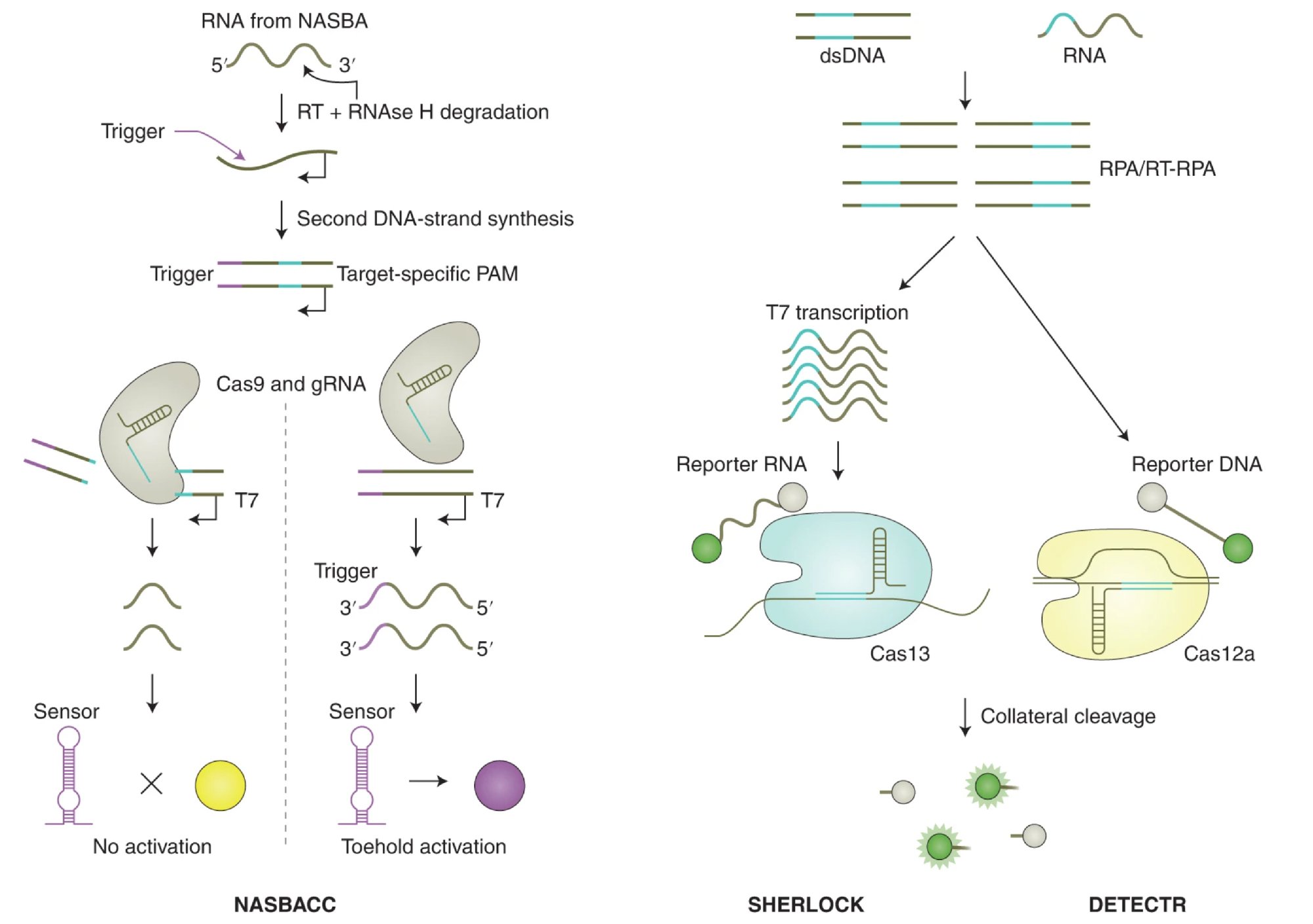
Fig. 2. Cleavage of assay reporters by Cas nucleases indicate the presence of RNA or DNA target sequences by SHERLOCK and DETECTR, respectively. (Credit: CRISPR-based diagnostics, Nature Biomedical Engineering)
Customization and Versatility
One of the significant advantages of CRISPR in assay development is its versatility. Because CRISPR can be programmed to target virtually any DNA or RNA sequence, it allows for the rapid development of custom assays tailored to specific applications. This is particularly valuable in fields like personalized medicine, where treatments and diagnostics need to be customized to individual patients' genetic profiles.
In drug discovery, CRISPR-based assays can be used to screen for the effects of genetic modifications or to identify potential drug targets. By creating assays that can directly measure the impact of specific gene edits on cellular functions, researchers can accelerate the identification of new therapeutic strategies. Furthermore, CRISPR’s ability to generate stable, cell-based assays through genome editing ensures more physiologically relevant models, enhancing the predictive power of preclinical studies.
EditCo Enables Assay Development with Effective CRISPR Solutions Delivered as Fast as You Need Them
Advanced CRISPR Reagents for High efficiency, Reliable Gene Knockouts

EditCo’s cutting-edge CRISPR reagents leverage an intelligent guide design strategy to deliver efficient and consistent gene knockouts. With intelligent guide design strategy, EditCo is able to simply and effectively knockout genes guaranteed with our Gene Knockout Kits or do high-throughput genomic screening with Arrayed gRNA Libraries, allowing for clear genotype-to-phenotype answers for many genes at scale and quickly.
Regardless of your throughput requirements, our solutions are designed to deliver accuracy and reliability in your experiments.
High-Throughput Screens with Arrayed gRNA Libraries
High-throughput screens are used to investigate genotype-phenotype relationships on a large scale. For each screen, a gRNA library is applied to cells to systematically knock out hundreds or thousands of genes. The phenotypic effects of each knockout can then be assessed. Loss-of-function screens can be used to identify gene function in entire biological pathways or disease states. They have also been critical to the process of drug discovery. For instance, a knockout that confers resistance or sensitivity to a drug may provide information about the drug’s target.
Type of Assay |
Pooled Screen |
Arrayed Screen |
| Cell Viability | ✔️ | ✔️ |
| Cell Proliferation | ✔️ | ✔️ |
| Drug Sensitivity | ✔️ | ✔️ |
| Synthetic Lethality | ✔️ | ✔️ |
| Response to Selection Pressure | ✔️ | ✔️ |
| FACS/Surface Marker Sorting | ✔️ | ✔️ |
| Cell Morphology | ✔️ | |
| Cell Differentiation, Migration, Phagocytosis, Clustering, etc. | ✔️ | |
| Intracellular Trafficking | ✔️ | |
| Time-course Studies | ✔️ | |
| Biochemical Assay Readouts (i.e. Colorimetric, Fluorescence, Luminescence) | ✔️ | |
| Protein-protein Interactions (i.e. Split Protein Assay, Homogenous Time Resolved Florescence) | ✔️ | |
| Analyte/Metabolite Detection (i.e. ELISA, Luciferase) | ✔️ |
CRISPR knockout cell pools with high KO frequencies can be used to screen small molecule libraries to identify potential therapeutic compounds. For instance, cell pools with a gene KO that induces a disease can be generated, and a library of compounds can be systematically applied. Resulting phenotypes can be assessed to determine if any of the compounds affected the disease state of the cells.
Streamlined Assay Development with Engineered Cells

EditCo’s KO and KI cell offerings provide researchers with precise reagents for assay development by enabling targeted modifications to specific genes. This allows for the creation of accurate cell models, facilitating the study of gene functions and interactions. These customized cells enhance the accuracy and relevance of assays, leading to more reliable and reproducible results.
Powered by our high-throughput CRISPR platform, EditCo engineered cells offer an affordable pathway to state-of-the-art knockouts and precision knock-ins, available in both pooled and clonal formats. EditCo offers a wide range of ATCC-supplied immortalized cell lines, iPS cells, and primary cells – OR – onboard your own cell lines!
Antibody Validation
All immunoassays utilize antibodies that are specific for their target proteins. For each assay, the specificity of an antibody to a particular protein must be validated to avoid non-specific results. To test antibody specificity, a cell pool can be generated with a knockout of the antibody’s protein target. Because less target protein is made in the pool, subsequent immunoblot or immunofluorescence analyses (which use the antibody to detect and visualize the protein) should have a proportionally lower signal. If this is observed, it is a good indication that the protein is indeed the antibody’s target.
Have more questions? Reach out to us!
